RNAEditor
A tool to detect editing sites from deep sequencing data
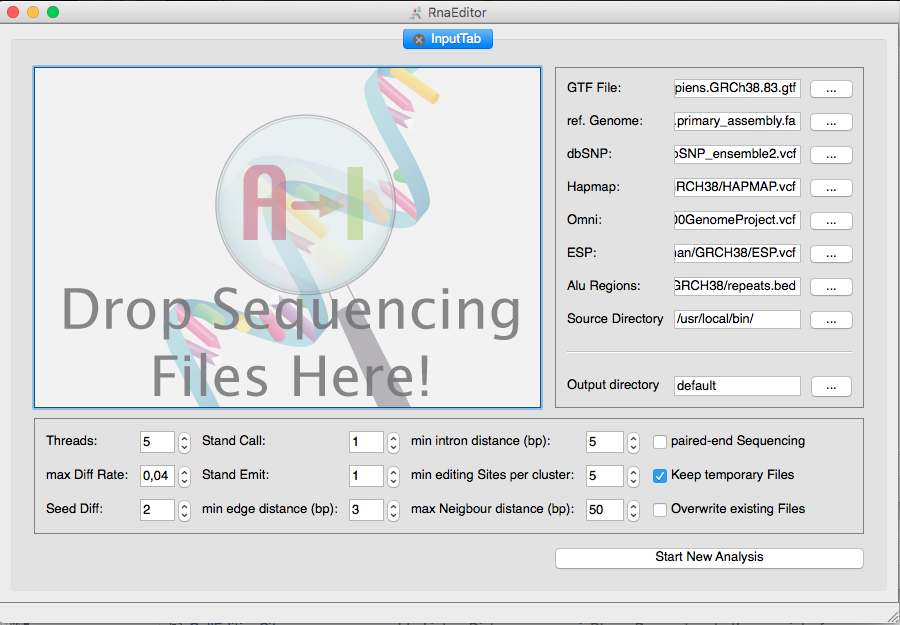
RNAEditor is an all-in-one tool to analyze RNA editing events from RNA sequencing (RNA-seq) data. Raw sequencing reads (.fastq) or mapped reads (.bam) can be used as an input data. RNAEditor maps the reads to the genome, calculates sequence variations, filters for “non-editing sites” and applies a cluster algorithm to detect “editing islands”.
References
If you use RNAEditor in your work, please cite the following articles!- John D, Weirick T, Dimmeler S, Uchida S.
RNAEditor: easy detection of RNA editing events and the introduction of editing islands.
Briefings in Bioinformatics 2016. [PMID: 27694136] - Stellos K, Gatsiou A, Stamatelopoulos K, Matic LP, John D, ... Zeiher A & Dimmeler S
Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation.
Nature Medicine 2016. [PMID: 27595325]
News of RNAEditor
- RNAEditor 1.0
23 June 2016
