RNAEditor Documentation
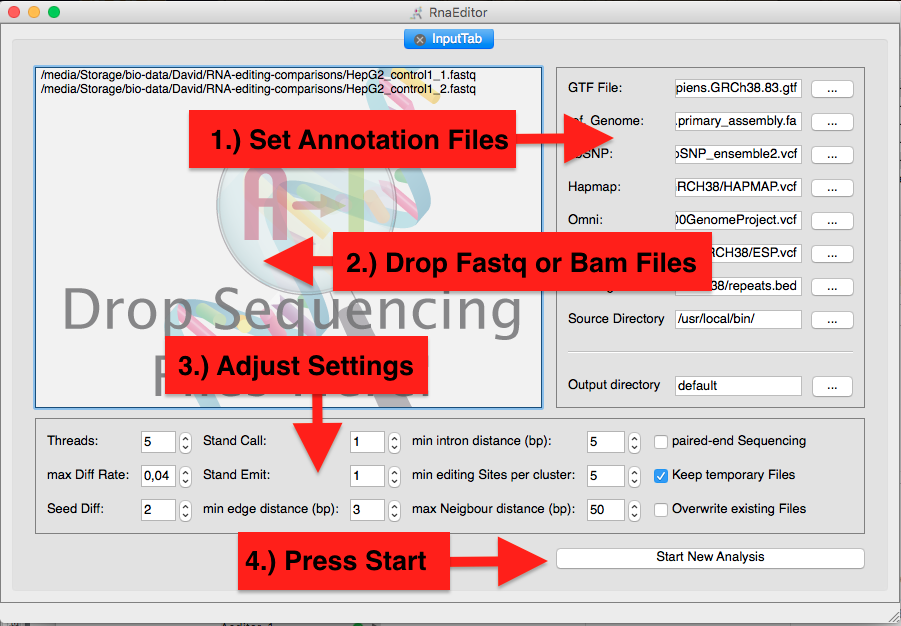
Starting Analysis
In the Input Tab you can Drop your sequencing files and adjust the parameters and the annotation files.
You can also drop a prepared configuration file from the annotation bundles.
Note: Before analyzing your data by RNAEditor, we highly recommend preprocessing steps like adapter and low quality trimming.
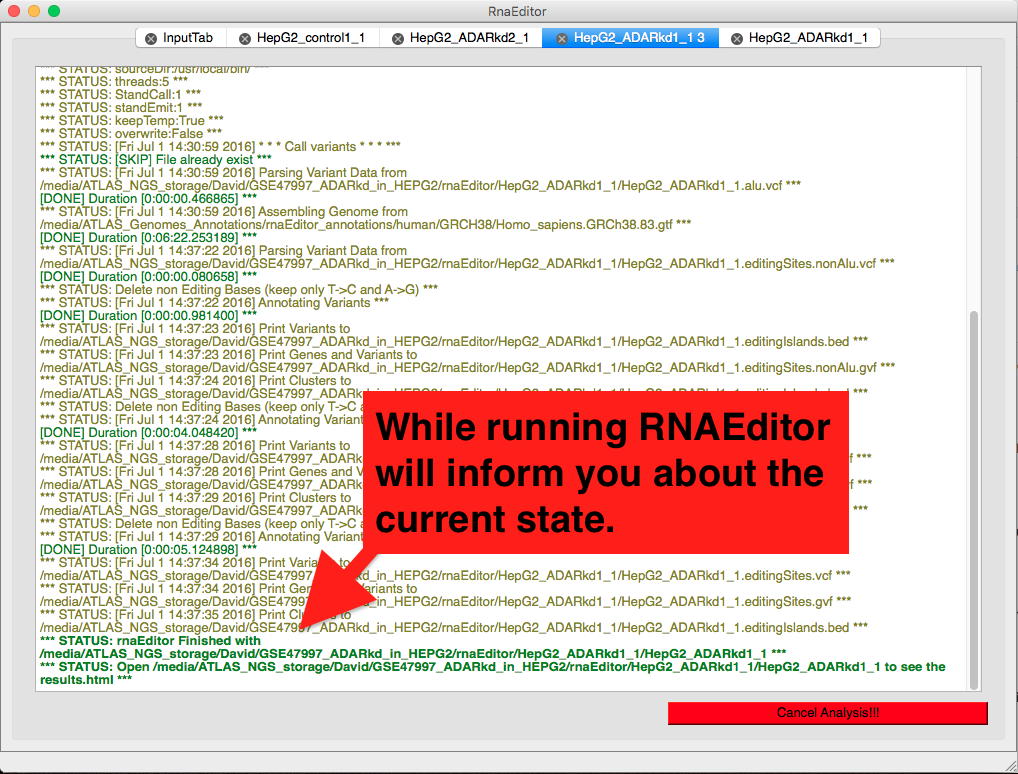
While Waiting
While RNAEditor provides detailed informnations on the current step of analysis.
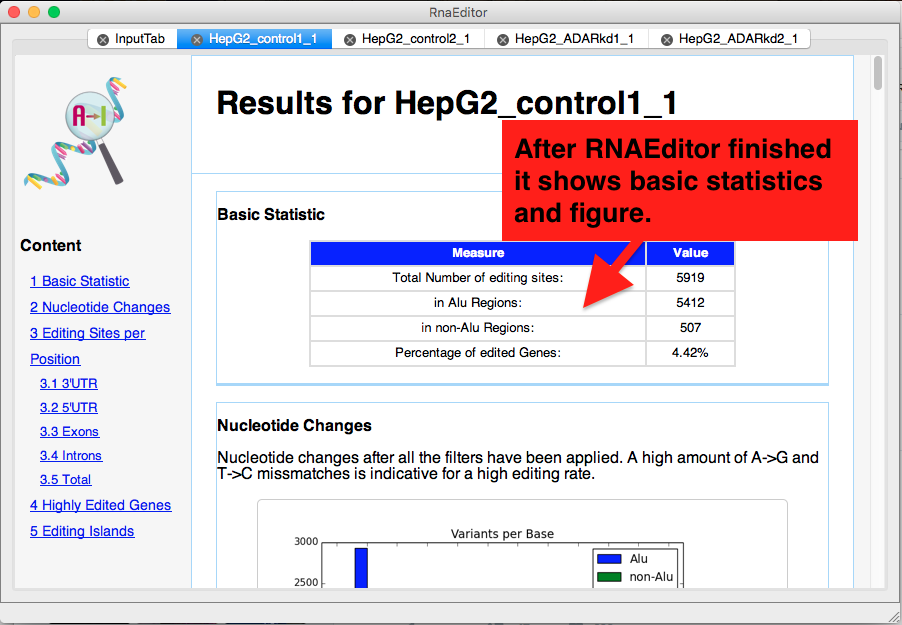
Once Finished
Once RNAEditor finished the analysis succesfully it will open the ResultTab which provides you with basic statistics about editing in your sample.
Information about total number of editing sites, highly edited genes, editing position and editing islands are shown.
Output Files
VCF File:
A standard vcf file with all the editing sites for further analysis.
Example: Sample.vcf
GVF File:
Gene Variation File hold additional informations for each editing site, like gene names, segments, number of total reads, number of edited reads and the ratio of editing.
Example: Sample.gvf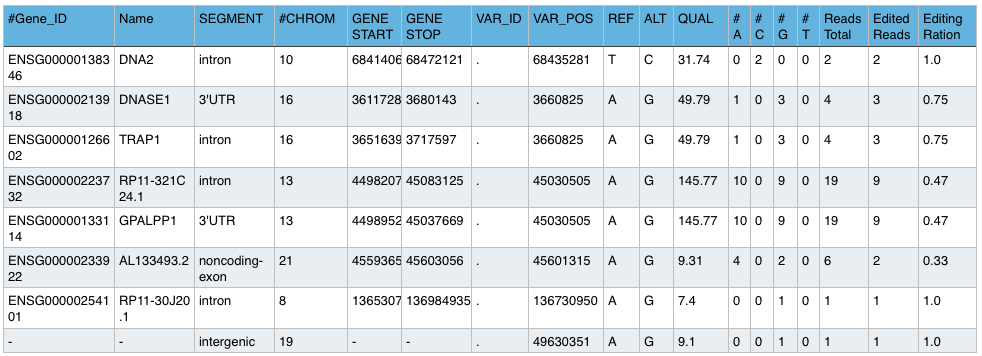
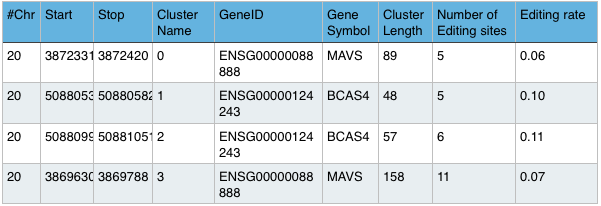
Summary File:
Shows the sum of editing sites per segment for each gene.
Example: Sample.summary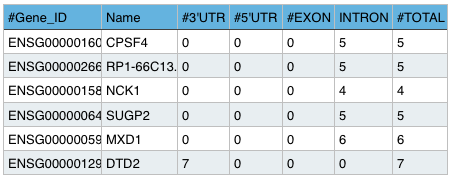
Editing Island File:
Example: Sample.clusters.bed